PSI4 API: Linking C++ and Python¶
psi4.core Module¶
C++ Innards of Psi4: Open-Source Quantum Chemistry
Functions¶
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
Runs the ADC propagator code, for excited states. |
|
Returns copy of the Matrix QCVariable key (case-insensitive); prefer |
Returns dictionary of all Matrix QCVariables; prefer |
|
|
Redirects output to |
|
Perform benchmark traverse of BLAS 1 routines. |
|
Perform benchmark traverse of BLAS 2 routines. |
|
Perform benchmark traverse of BLAS 3 routines. |
|
Perform benchmark of PSIO disk performance. |
|
Perform benchmark of psi integrals (of libmints type). |
|
Perform benchmark of common double floating point operations including most of cmath. |
|
Runs the code to compute coupled cluster density matrices. |
|
Runs the coupled cluster energy code. |
|
Runs the equation of motion coupled cluster code for excited states. |
|
Runs the code to generate the similarity transformed Hamiltonian. |
|
Runs the coupled cluster lambda equations code. |
|
Runs the coupled cluster response theory code. |
|
Runs cctransort that transforms and reorders integrals for use in the coupled cluster codes. |
|
Runs the coupled cluster (T) energy code. |
|
Remove scratch files. |
Reset options to clean state. |
|
Reinitialize timers for independent |
|
Empties all double and Matrix QCVariables that have been set in global memory. |
|
Closes the output file. |
|
|
Runs the density cumulant (functional) theory code. |
|
Removes the Matrix QCVariable key (case-insensitive); prefer |
|
Removes the double QCVariable key (case-insensitive); prefer |
|
Removes scalar or array QCVariable key from global memory if present. |
|
Runs the determinant-based configuration interaction code. |
|
Runs the DF-MP2 code. |
|
Runs the density-fitted orbital optimized CC codes. |
|
Runs the DLPNO codes. |
|
Runs the CheMPS2 interface DMRG code. |
|
Returns the multiplication of two matrices A and B, with options to transpose each beforehand |
|
Write integrals to file in FCIDUMP format |
|
Called upon psi4 module exit to closes timers and I/O. |
Flushes the output file. |
|
|
Runs the FNO-CCSD(T)/QCISD(T)/MP4/CEPA energy code |
Returns the currently active molecule object. |
|
|
Deprecated since version 1.4. |
Deprecated since version 1.4. |
|
Deprecated since version 1.4. |
|
Returns the path to shared text resources, |
|
|
Return keyword key value at global (all-module) scope. |
Returns a list of all global options. |
|
Deprecated since version 1.2. |
|
Returns the global gradient as a (nat, 3) |
|
Returns the currently active legacy molecule object. |
|
|
Return keyword key value at module scope. |
Returns the amount of memory available to Psi (in bytes). |
|
Returns the number of threads to use in SMP parallel computations. |
|
|
Return keyword key value used by module. |
Get options |
|
Returns output file name (stem + suffix, no directory). |
|
|
Deprecated since version 1.4. |
Deprecated since version 1.4. |
|
|
Returns the prefix to use for writing files for external programs. |
Deprecated since version 1.4. |
|
|
Is the Matrix QCVariable key (case-insensitive) set? Prefer |
Whether keyword key value has been touched at global (all-module) scope. |
|
|
Whether keyword key value has been touched at module scope. |
|
Whether keyword key value has been touched or is default. |
|
Is the double QCVariable key (case-insensitive) set? Prefer |
|
Whether scalar or array QCVariable key has been set in global memory. |
Called upon psi4 module import to initialize timers, singletons, and I/O. |
|
Returns the current legacy_wavefunction object from the most recent computation. |
|
|
Runs the MCSCF code, (N.B. |
|
Generates an input for Kallay's MRCC code. |
|
Reads in the density matrices from Kallay's MRCC code. |
|
Runs the orbital optimized CC codes. |
Cleans up the optimizer's scratch files. |
|
|
Whether keyword key is a valid keyword for module. |
|
Get dictionary of whether options of module have changed. |
|
Runs the geometry optimization code. |
Returns the name of the output file. |
|
|
Call the plugin of name arg0. |
|
Close the plugin of name arg0. |
Close all open plugins. |
|
|
Load the plugin of name arg0. |
Sets up the options library to return options pertaining to the module or plugin name (e.g. |
|
Prints the currently set global (all modules) options to the output file. |
|
Prints the currently set options (to the output file) for the current module. |
|
|
Prints a string (using sprintf-like notation) to the output file. |
Prints to output file all QCVariables that have been set in global memory. |
|
Returns the location of the source code. |
|
|
Runs the multireference coupled cluster code. |
Reopens the output file. |
|
Clear the touched status for keyword key at global (all-module) scope. |
|
|
Clear the touched status for keyword key at module scope. |
|
Runs the GDMA interface code. |
|
Runs the symmetry adapted perturbation theory code. |
|
Returns the double QCVariable key (case-insensitive); prefer |
Returns dictionary of all double QCVariables; prefer |
|
|
New Scatter function. |
|
Run scfgrad, which is a specialized DF-SCF gradient program. |
|
Run scfhess, which is a specialized DF-SCF hessian program. |
|
Activates a previously defined molecule in global memory so next computations use it. |
|
Sets the requested (case-insensitive) Matrix QCVariable; prefer |
|
Sets the path to shared text resources, |
|
Overloaded function. |
|
This is a fairly hacky way to get around EXTERN issues. |
|
Deprecated since version 1.2. |
|
Assigns the global gradient to the values in the (nat, 3) Matrix argument. |
|
Activates a previously defined molecule in global memory so next computations use it. |
Sets the current legacy_wavefunction object from the most recent computation. |
|
|
Overloaded function. |
|
Sets value to Python keyword key scoped only to a single module. |
|
Sets the memory available to Psi (in bytes); prefer |
|
Sets the number of threads to use in SMP parallel computations. |
|
Overloaded function. |
Deprecated since version 1.4. |
|
|
Sets the double QCVariable key (case-insensitive); prefer |
|
Sets scalar or array QCVariable key to val in global memory. |
|
Stop timer with label. |
|
Start timer with label. |
|
Returns the multiplication of three matrices, with options to transpose each beforehand. |
|
Start module-level timer. |
|
Stop module-level timer. |
|
Return copy of scalar or array QCVariable key from global memory. |
|
Return all scalar or array QCVariables from global memory. |
|
Deprecated since version 1.4. |
Classes¶
Computes angular momentum integrals |
|
docstring |
|
docstring |
|
Contains basis set information |
|
docstring |
|
Performs Boys orbital localization |
|
Specialized Wavefunction used by the ccenergy, cceom, ccgradient, etc. |
|
docstring |
|
docstring |
|
docstring |
|
Cartesian displacement SALC |
|
Class for generating symmetry adapted linear combinations of Cartesian displacements |
|
Contains the character table of the point group |
|
docstring |
|
Provides a correlation table between two point groups |
|
docstring |
|
A density-fitted second-order Electron Propagator Wavefunction. |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
Computes gradients of wavefunctions |
|
Members: |
|
Defines ordering of eigenvalues after diagonalization |
|
Initializes and defines Dimension Objects |
|
Computes dipole integrals |
|
docstring |
|
docstring |
|
docstring |
|
Computes normal two electron repulsion integrals |
|
docstring |
|
ESPPropCalc gives access to routines calculating the ESP on a grid |
|
Computes electric field integrals |
|
Computes electrostatic integrals |
|
Stores external potential field, computes external potential matrix |
|
Extracts information from a wavefunction object, and writes it to an FCHK file |
|
docstring |
|
A Fragment-SAPT Wavefunction |
|
docstring |
|
docstring |
|
Fragment activation status |
|
docstring |
|
Class containing information about basis functions |
|
0 if Cartesian, 1 if Pure |
|
The units used to define the geometry |
|
docstring |
|
docstring |
|
PSIOManager is a class designed to be used as a static object to track all PSIO operations in a given PSI4 computation |
|
Class handling vectors with integer values |
|
Computes integrals |
|
IntegralTransform transforms one- and two-electron integrals within general spaces |
|
An irreducible representation of the point group |
|
docstring |
|
Computes kinetic integrals |
|
docstring |
|
docstring |
|
Class containing orbital localization procedures |
|
Defines orbital spaces in which to transform integrals |
|
Writes the MOs |
|
Class for creating and manipulating matrices |
|
Creates Matrix objects |
|
docstring |
|
Computes integrals |
|
Writes wavefunction information in molden format |
|
docstring |
|
Class to store the elements, coordinates, fragmentation pattern, basis sets, charge, multiplicity, etc. |
|
Computes arbitrary-order multipole integrals |
|
docstring |
|
The Natural Bond Orbital Writer |
|
Computes nabla integrals |
|
docstring |
|
Basis class for all one-electron integrals |
|
docstring |
|
Contains information about the orbitals |
|
Computes overlap integrals |
|
Performs Pipek-Mezey orbital localization |
|
Handles symmetry transformations |
|
docstring |
|
Contains information about the point group |
|
Computes potential integrals |
|
May be Normalized or Unnormalized |
|
docstring |
|
Computes pseudospectral integrals |
|
Return status. |
|
Computes quadrupole integrals |
|
docstring |
|
docstring |
|
docstring |
|
docstring |
|
An SOBasis object describes the transformation from an atomic orbital basis to a symmetry orbital basis. |
|
docstring |
|
Component of a Cartesian displacement SALC in the basis of atomic displacements. |
|
The layout of the matrix for saving |
|
Slicing for Matrix and Vector objects |
|
docstring |
|
Class to provide a 3 by 3 matrix representation of a symmetry operation, such as a rotation or reflection. |
|
docstring |
|
Three center overlap integrals |
|
Computes traceless quadrupole integrals |
|
Two body integral base class |
|
Computes two-electron repulsion integrals |
|
docstring |
|
docstring |
|
docstring |
|
Class for creating and manipulating vectors |
|
Class for vectors of length three, often Cartesian coordinate vectors, and their common operations |
|
docstring |
|
docstring |
Class Inheritance Diagram¶

psi4.driver Package¶
Functions¶
|
Function to set molecule object mol as the current active molecule. |
|
Get the nth ancestor of a directory. |
|
Function to print text to output file in a banner of minimum width width and minimum three-line height for type = 1 or one-line height for type = 2. |
|
For PsiAPI mode, forms a basis specification function from block and associates it with keyword key under handle name. |
|
Function to define a multistage energy method from combinations of basis set extrapolations and delta corrections and condense the components into a minimum number of calculations. |
|
Ensures that a IWL file has been written based on input SCF type. |
|
|
|
Function to compare two FCIDUMP files. |
|
Function to move file out of scratch with correct naming convention. |
|
Function to move file into scratch with correct naming convention. |
|
|
|
Evaluate properties on a grid and generate cube files. |
Function to dynamically add extra members to the core.Molecule class. |
|
|
|
|
Function to compute the single-point electronic energy. |
|
Function to write wavefunction information in wfn to filename in Gaussian FCHK format. |
|
Load FCHK file into a string |
|
Save integrals to file in FCIDUMP format as defined in Comp. |
|
Function to read in a FCIDUMP file. |
|
Find list of approximate (within max_distance) matches to string seq1 among options. |
|
Computes free-atom volumes using MBIS density partitioning. |
|
Function to compute harmonic vibrational frequencies. |
|
Function to compute harmonic vibrational frequencies. |
|
Function to compute harmonic vibrational frequencies. |
|
Function to use wavefunction information in wfn and, if specified, additional commands in filename to run GDMA analysis. |
|
Function to create a molecule object of name name from the geometry in string geom. |
Function to return the total memory allocation. |
|
|
Function complementary to |
|
Function complementary to |
|
Run IPIBroker to connect to i-pi |
|
|
|
Compute the Levenshtein distance between two strings. |
|
Begin functioning as an MDI engine |
|
put a message string into a box for extra attention |
|
Function to write wavefunction information in wfn to filename in molden format. |
|
Function to redefine __getattr__ method of molecule class. |
|
Function to redefine __setattr__ method of molecule class. |
|
Evaluate one-electron properties. |
|
Function to perform a geometry optimization. |
|
Function to perform a geometry optimization. |
|
|
|
Passes multiline string block to PCMSolver parser. |
|
Function to print stuff to standard error stream. |
|
Function to print stuff to standard output stream. |
|
Function to preprocess raw input, the text of the input file, then parse it, validate it for format, and convert it into legitimate Python. |
|
Function to compute various properties. |
|
Function to compute various properties. |
|
Function to return name in coded form, stripped of characters that confuse filenames, characters into lowercase, |
|
Function serving as helper to SCF, choosing whether to cast up or just run SCF with a standard guess. |
|
Builds the correct (R/U/RO/CU HF/KS) wavefunction from the provided information, sets relevant auxiliary basis sets on it, and prepares any empirical dispersion. |
|
Function to reset the total memory allocation. |
|
Sets Psi4 module options from a module specification and input dictionary. |
|
Sets Psi4 options from an input dictionary. |
|
|
|
Function to perform analysis of a hessian or hessian block, specifically. |
|
Prepare multi-line string with one-particle eigenvalues to be written to the FCIDUMP file. |
Classes¶
|
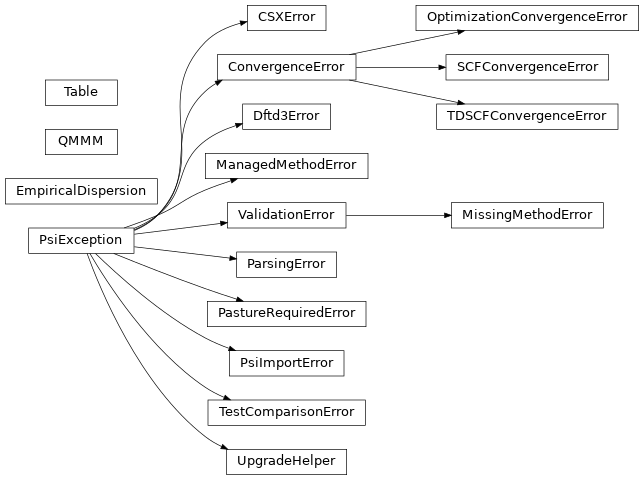
Error called when CSX generation fails. |
|
Error called for problems with converging an iterative method. |
|
|
|
Lightweight unification of empirical dispersion calculation modes. |
|
|
|
Error called when method not available. |
Error called for problems with geometry optimizer. |
|
|
Error called for problems parsing a text file. |
|
Error called when the specified value of option requires some module(s) from Psi4Pasture, but could not be imported. |
Error class for Psi. |
|
|
Error called for problems import python dependencies. |
|
|
|
Error called for problems with SCF iterations. |
|
Error called for problems with TDSCF iterations. |
|
Class defining a flexible Table object for storing data. |
|
Error called when a test case fails due to a failed compare_values() call. |
|
Error called on previously valid syntax that now isn't and a simple syntax transition is possible. |
|
Error called for problems with the input file. |
Class Inheritance Diagram¶